Digital Lab Notebook
Comprehensive documentation of our experimental protocols, observations, and results for the POP2 cloning and expression project.
Lab Notebook
Project Overview & Purpose
This digital lab notebook documents our experimental protocols, observations, and results for the POP2 cloning and expression project. All procedures are presented in chronological order with specific details to ensure reproducibility. This document serves as an accurate record of our laboratory work conducted in February-March 2025.
Protocols Overview
Our research utilized several key protocols for protein expression and analysis. Below is a summary of the main protocols used in this project.
E. coli Transformation
Standard protocol for transforming E. coli with plasmid DNA using heat shock method.
Plasmid Mini-Prep
Protocol for isolating plasmid DNA from bacterial cultures using ZymoPURE kit.
Restriction Enzyme Digest
Protocol for digesting plasmid DNA with restriction enzymes for cloning.
Expression Optimization
Protocol for optimizing protein expression conditions in E. coli.
Lab Equipment & Reagents
Equipment Used
- Centrifuges:
- Eppendorf Centrifuge 5910R (45,000g max)
- Eppendorf Centrifuge 5418
- Incubators:
- Mere Instruments 747R Shaking Incubator
- 10L Shaking Incubator VWR
- Spectrophotometers:
- Thermo Scientific NanoDrop One
- Genesys 10UV Spectrophotometer
- Gel Documentation:
- BioRad Gel Doc Easy Imager
- Power Supply:
- BioRad PowerPac Basic
Media and Reagents
- LB Medium:
- Standard composition: 10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl
- 2×YT Medium (BC404):
- Prepared on 1/16/25
- Ampicillin:
- 1000× stock
Initial Experiments
Experiment Timeline
Plasmid Preparation & Transformation
February 25, 2025
Reconstitution of lyophilized plasmid and transformation into E. coli cells
Colony Assessment & Culture Setup
February 26, 2025
Evaluation of transformation success and preparation of overnight cultures
Plasmid Isolation (Mini-Prep)
February 27, 2025
Extraction and purification of plasmid DNA from bacterial cultures
Protocol 1: Plasmid Reconstitution
Purpose: Reconstitution of lyophilized plasmid DNA (pPOP2-E and pPOP2-Y) with TE buffer to prepare for transformation.
Materials:
- Lyophilized plasmid (pPOP2-E and pPOP2-Y)
- TE buffer
- -20°C freezer
Procedure:
- Add TE buffer to the lyophilized plasmid (volume not measured).
- Gently pipette up and down to ensure complete reconstitution.
- Allow to sit at room temperature for approximately 30-60 minutes.
- Store the reconstituted plasmid stock at −20°C.
Protocol 2: E. coli Transformation
Purpose: Introduction of plasmid DNA into competent E. coli cells using heat shock method to create transformed bacteria.
Materials:
- Competent cells: pTwist Amp High Copy (2221 bp) with pMB1 origin of replication
- Plasmids: pPOP2-E (pTwist Amp for E. coli) and pPOP2-Y (pRS423-GAL for yeast)
- SOC media (Super Optimal Broth with Catabolite Repression)
- LB + Ampicillin plates
- Ice bucket
- 42°C water bath
- 37°C shaking incubator
- Sterile 1.5 mL microcentrifuge tubes
- Sterile spreader or glass beads
Procedure:
- Thaw competent cells on ice for approximately 30-60 minutes.
- Label sterile 1.5 mL microcentrifuge tubes for each transformation.
- Combine 20 µL of competent cells with 1 µL plasmid (pPOP2-E or pPOP2-Y).
- Incubate the mixture on ice for 30 minutes.
- Heat-shock at 42°C for exactly 30 seconds.
- Add 219 µL SOC media to bring total volume to 250 µL.
- Incubate at 37°C with shaking for 1 hour.
- Plate 125 µL on LB + Amp plates.
- Incubate overnight (16-18 hours) at 37°C.
- Hundreds of colonies were observed but exact counts were not recorded.
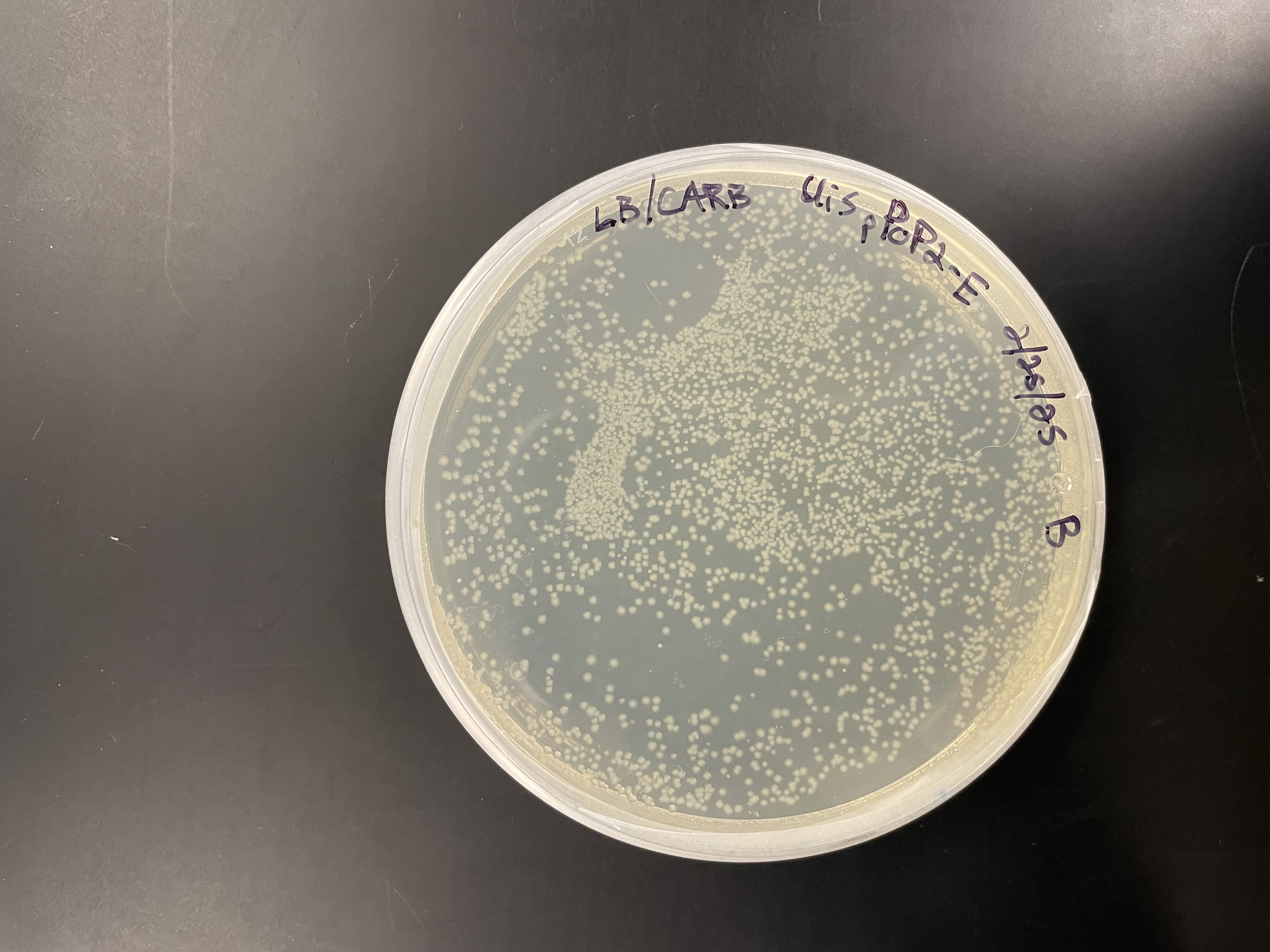
E. coli colonies on LB + Ampicillin plates after transformation
Protocol 3: Colony Assessment & Culture Setup
Purpose: Evaluation of transformation success and preparation of liquid cultures from selected colonies for subsequent plasmid isolation.
Materials:
- Transformation plates from previous day
- Sterile 15 mL culture tubes
- LB broth
- Ampicillin (1000× stock)
- Sterile pipette tips
- Parafilm
- 37°C shaking incubator
- 4°C refrigerator
Procedure:
- Colony Assessment (10:30 am):
- Remove plates from incubator and examine for colony growth.
- Hundreds of colonies were observed but exact counts were not recorded.
- Seal plates with parafilm and store at 4°C.
- Overnight Culture Preparation (6:00 pm):
- Prepare 4 culture tubes (15 mL capacity) containing 5 mL LB + Ampicillin each.
- For each tube: Add 5 µL of 1000× Amp stock to 5 mL LB.
- Label tubes A, B, C, and D with plasmid name and date.
- Using a sterile pipette tip, pick a single, well-isolated colony from each transformation plate.
- Inoculate each tube with a single colony.
- Incubate at 37°C overnight (12-16 hours) with shaking.
Protocol 4: Plasmid Mini-Prep
Purpose: Extraction and purification of plasmid DNA from bacterial cultures using the ZymoPURE mini prep kit for downstream applications.
Materials:
1. Sample Preparation:
- Label tubes for each sample
- Obtain one spin column and one collection tube per sample
2. Cell Harvesting:
- Transfer 1.5 mL from each overnight culture to corresponding 1.5 mL tube
- Centrifuge at 14,000 g for 30 seconds to pellet cells
- Discard supernatant by pipetting
- Repeat with additional 1.5 mL from the same culture
3. Cell Lysis:
- Add 250 µL P1 buffer (red) to each pellet
- Resuspend completely by pipetting
- Add 250 µL P2 buffer (green) to each tube
- Gently invert tubes exactly 8 times
- Incubate at room temperature for exactly 3 minutes
- Add 250 µL P3 buffer (yellow) to each tube
- Gently invert tubes exactly 5 times until a white precipitate forms
4. DNA Purification:
- Centrifuge at maximum speed (14,000 g) for 5 minutes
- Transfer supernatant to spin column
- Wash column with provided buffers
- Elute DNA with 25 µL Elution Buffer
- Store isolated plasmid at -20°C for long-term storage