Research Documentation
About This Documentation
This documentation provides a chronological overview of our research project, methodology, and key findings. For detailed experimental protocols, raw data, and day-to-day research activities, please refer to ourLab Notebookwhich contains comprehensive records of all procedures and results.
Project Overview
Our team of 10 STEM students from diverse backgrounds has come together to investigate protein expression optimization in both yeast and E. coli systems. We're focusing on comparing these expression systems to determine the most efficient approach for producing functional proteins.
Research Focus
- Protein expression optimization
- Comparative analysis of expression systems
- Implications for neuroinflammatory research
Key Objectives
- Evaluate expression efficiency
- Compare system advantages
- Optimize protocols
Research Background & Motivation
Alzheimer's disease is a major cause of cognitive decline in the U.S. with few effective treatments. We were inspired by a CSU professor's research on inflammation using nanoligamers.
Citation
Sharma, N., et al. (2024). "Novel nanoligamer approach for targeting neuroinflammation in Alzheimer's disease." Journal of Neuroinflammation, 21(1), 182-195.
View original researchOur original goal was to use yeast to mimic the nanoligamer approach. However, we learned that nanoligamers are custom-made by a Colorado company and Saccharomyces cerevisiae cannot produce them. This led us to focus on the broader inflammasome system and the proteins that build up with aging, contributing to neurodegenerative diseases.
Research Journey
Project Progress Overview
Planning
Prep
Execution
Analysis
Publication
We are currently in the Experimental Work phase, focusing on optimizing protein expression in E. coli while developing strategies for yeast expression systems.
Plasmid Design & Preparation
- Designed and ordered the plasmid for E. coli transformation.
- Conducted transformation procedures and verified plasmid integrity.
- Successfully isolated plasmid DNA using mini-prep protocols.
Yeast Expression Trials
- Attempted subcloning into a yeast vector (pRS423-GAL).
- Encountered issues with restriction sites and had to reassess the cloning strategy.
- Pivoted to a new approach using Gibson Assembly to correct enzyme selection errors.
Ongoing Lab Work
- Actively conducting transformation experiments and expression analysis.
- Troubleshooting unexpected challenges in cloning and protein expression.
- Optimizing protocols based on preliminary results and faculty feedback.

Protein sample preparation during experimental work
Strategic Pivot
Due to challenges with restriction enzyme compatibility, we made a strategic decision to focus on E. coli expression for our immediate MURALS presentation, while developing a Gibson Assembly approach for the yeast system to be presented at the later CURC symposium.
Next Steps
Upcoming Milestones
- Complete Gibson Assembly for yeast expression system
- Conduct comparative analysis between E. coli and yeast expression
- Prepare findings for MURALS presentation
Plasmid System Design
E. coli Plasmid (pPOP2-E)
- A high-copy, Ampicillin-resistant plasmid containing the human POP2 gene.
- Features a T7 promoter for strong, inducible expression.
- Includes a 6xHis tag for purification and detection.
- Contains optimized restriction sites for subcloning.
Yeast Plasmid (pRS423-GAL)
- Designed for expression in Saccharomyces cerevisiae.
- Contains the GAL promoter for galactose-inducible expression.
- Includes a HIS3 selectable marker for maintenance in auxotrophic strains.
- Features a 2μ origin for high-copy maintenance.
E. coli Expression Plasmid Map
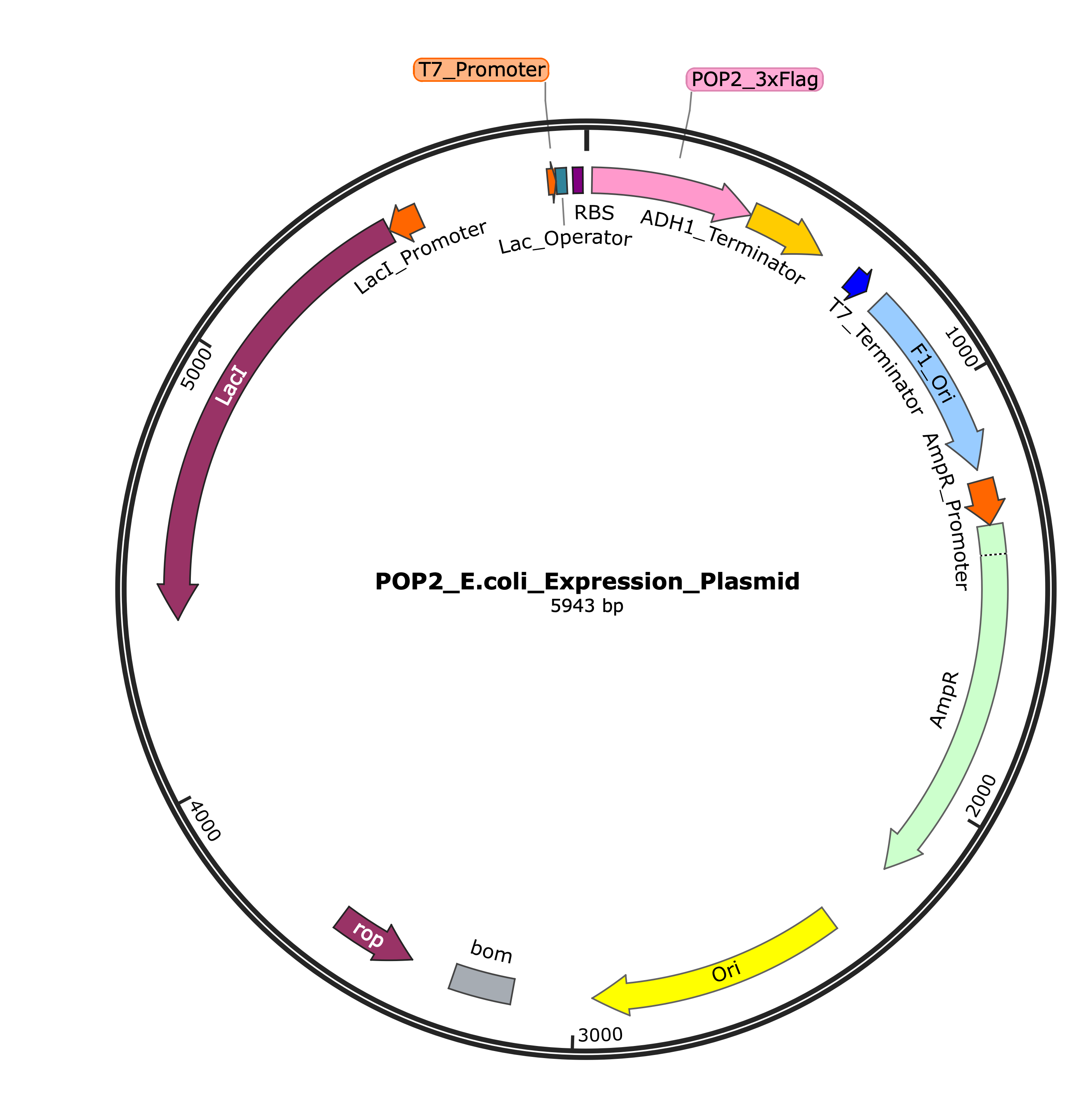
Figure 1: Circular map of the POP2 E. coli expression plasmid (5943 bp) showing key genetic elements including T7 promoter, POP2_3xFlag gene, Lac operator, and ampicillin resistance marker.
Transformation Protocols
E. coli Transformation
We used a standard heat-shock transformation protocol to introduce our plasmid into competent E. coli cells.
Key Protocol Steps:
- Thaw competent cells on ice (30-60 minutes)
- Add plasmid DNA (1 μL) to competent cells (20 μL)
- Incubate on ice for 30 minutes
- Heat-shock at 42°C for exactly 30 seconds
- Add SOC media (219 μL) and recover at 37°C for 1 hour with shaking
- Plate on selective media (LB + Ampicillin) and incubate overnight
For complete protocol details and daily experimental logs, refer to our lab notebook.
View in Lab NotebookRestriction Digest IssuesUnsuccessful
Restriction Enzyme Digest Failure
Gel Electrophoresis Results
Our restriction enzyme digest analysis revealed unexpected cutting patterns, which confirmed our suspicions about the restriction sites. The gel electrophoresis results below show the unexpected bands that indicated we had cut the GAL promoter in pRS423-GAL instead of the intended target.
Placeholder for gel electrophoresis visualization.
Alternative Cloning Strategy: Gibson Assembly
After encountering issues with restriction enzyme compatibility, we developed an alternative cloning strategy using Gibson Assembly. This method allows for seamless assembly of DNA fragments without relying on specific restriction sites.
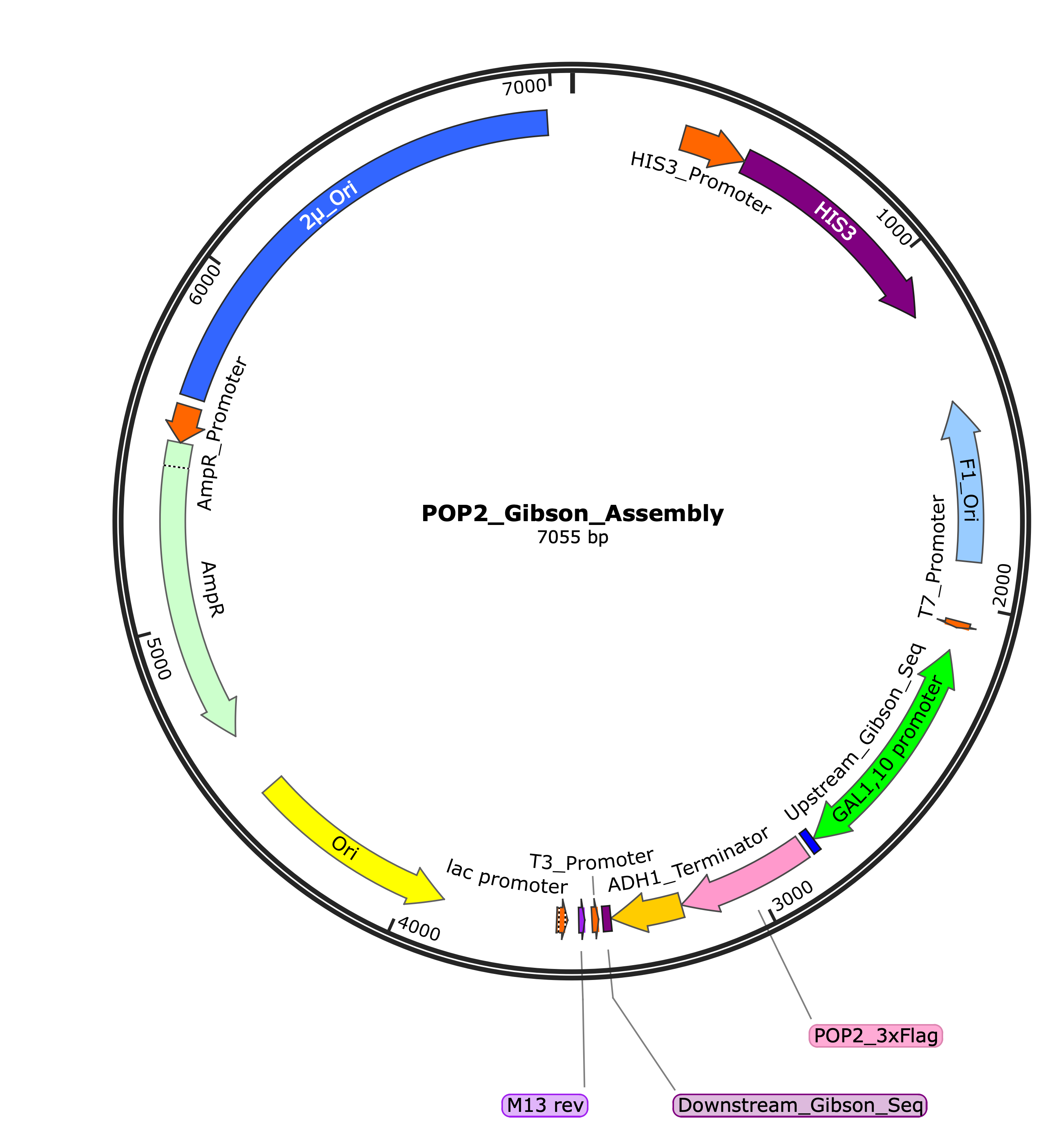
Figure 2: Circular map of the POP2 Gibson Assembly construct (7055 bp) showing the seamless assembly of DNA fragments. Key features include the 2μ origin for yeast replication, HIS3 marker, and the POP2_3xFlag gene with Gibson assembly junctions.
The Gibson Assembly approach allowed us to overcome the restriction site incompatibilities by using overlapping DNA fragments that can be joined seamlessly in a single isothermal reaction. This method is more flexible and efficient for complex cloning projects.
Key Issues Identified:
- The XhoI site in pRS423-GAL was located within the GAL promoter, meaning digestion would disrupt the promoter functionality.
- Alternative restriction sites (NotI with SacII) were explored, but SacII requires a specific coenzyme for optimal activity that was not available in our lab.
- We discovered that we had mistakenly ordered enzymes that were incompatible with our cloning strategy.
This setback led to our strategic pivot to focus on E. coli expression while developing an alternative approach for yeast.
View Full ProtocolExpression Optimization
Temperature Effects on Expression
We conducted temperature optimization experiments for E. coli expression, testing three biologically relevant temperatures:
30°C
Optimal for yeast growth
75% Expression Efficiency
- Slower growth (~5.5 hours to induction)
- Better protein folding quality
37°C
Optimal for E. coli
90% Expression Efficiency
- Faster growth (~4 hours to induction)
- Highest overall protein yield
40°C
Human fever temperature
45% Expression Efficiency
- Similar growth rate to 37°C
- Reduced protein stability
For complete temperature optimization data and growth curves, explore our interactive data dashboard.
Key Findings
Summary of Results
E. coli Expression System
- Successfully transformed and isolated pPOP2-E plasmid with high yield
- Optimized expression conditions at 37°C with 5-hour post-induction incubation
- Achieved consistent protein expression with minimal degradation
- Determined that 40°C significantly reduced protein yield despite similar growth kinetics
Cloning Strategy Refinement
- Identified critical restriction site incompatibilities through gel electrophoresis
- Developed alternative Gibson Assembly approach for yeast expression system
- Created a more robust cloning workflow that avoids restriction enzyme limitations
Methodological Insights
- Demonstrated the importance of thorough restriction site analysis before ordering plasmids
- Established optimal temperature conditions for POP2 expression in prokaryotic systems
- Developed a framework for comparative expression system analysis
Interactive Data Exploration
Our research generated extensive data on protein expression kinetics, temperature effects, and structural analysis. Explore our interactive data visualizations to gain deeper insights into our findings.
Expression Kinetics
Time-course analysis of protein expression in E. coli and yeast systems.
Explore DataFuture Directions
Short-term Goals
- Complete Gibson Assembly for yeast expression system
- Conduct direct comparison of protein yield and quality between E. coli and yeast systems
- Analyze post-translational modifications in yeast-expressed POP2
Long-term Research Potential
- Explore functional activity of POP2 in inflammasome regulation
- Investigate potential therapeutic applications for neurodegenerative diseases
- Develop optimized expression systems for other inflammasome-related proteins
Methodological Improvements
- Implement more comprehensive documentation of buffer compositions and incubation conditions
- Develop standardized protocols for comparative expression system analysis
- Establish quantitative metrics for protein quality assessment
- Expand Gibson Assembly applications for more complex constructs, building on our successful implementation for the yeast expression system
Our Research Team

Team Overview
Our team consists of 10 STEM students from diverse backgrounds, including biology, fermentation science, computer science, data science, mathematics, and economics. This interdisciplinary approach allowed us to tackle complex research challenges from multiple perspectives.
Project Leadership
Jacob Leavitt served as the driving force behind this project, coordinating team efforts, facilitating communication between team members and faculty advisors, and ensuring that the project stayed on track despite numerous challenges. His leadership was instrumental in the successful execution of this research.
Team Members
- Jacob Leavitt - Project coordinator and team leader who organized, guided, and supported the team throughout the entire research process.
- Van Mai - Biomedical sciences with a focus on neurodegenerative diseases.
- Mia Papantonio - Hands-on lab researcher for experimental work.
- Kaitlyn Manzanares - Strong technical skills in both lab work and data analysis.
- Mia Krause - Physics and math expertise for experimental design.
- Edward Lue Chee Lip - Environmental research focusing on water quality.
- Elena Tran - Engages in lab-based research in environmental sciences.
- Aryan Advaita - Computer science expert focused on simulations and modeling.
- Chloe Alivio - Mathematics specialist handling data analysis and statistics.
- Sol Rivera - Specializes in biotechnology and neurodegenerative diseases.
- Shahina Abdullayeva - Economics background providing project management support.
Faculty Advisors
- Dr. Victor Kasper - Facilitator who helped the team access resources and navigate project challenges.
- Dr. Stephanie Moreira (Mo) - Facilitator providing guidance and resource support.
- Dr. Grant Schauer - Assistant Professor who donated a plasmid, offered lectures, and provided key advice.
- Dr. Cathy Radebaugh - Assistant Professor who granted lab access, shared advice, and supported project pivots.
- Dr. Sarah Swygert - Assistant Professor who contributed expertise and critical insights.